
phylogeographic data aggregation and repurposing
First time users must create an account to access the phylogatR database. Sign up instructions can be found here.
This material is based upon work supported by the National Science Foundation under grant numbers 1910623 and 1911293
About
phylogatR
PhylogatR brings together genetic data with georeferenced specimen records that are analysis-ready. Analysis of these data can be conducted on the Ohio Supercomputer using R scripts or R Shiny apps provided by the phylogatR team. PhylogatR supports both professional biologists and educational modules intended for secondary or college classes.
How phylogatR Works
Patterns of genetic diversity within species contain information about the history of the species, including how it responded to historical climate change and the degree of isolation of its populations. For this reason, thousands of scientific projects have been funded by federal agencies with the goal of investigating the genetic diversity within a particular focal species. The data from these efforts, including the genetic data collected from a given specimen and the physical location from which that specimen was collected, are a valuable resource and typically are made available to other researchers upon publication of the study. However, since different types of data are stored in different databases, it can be difficult to access and assemble these data for reanalysis. PhylogatR is a database and web application that (i) aggregates genetic sequence data from GenBank and geographic occurrence data from the Global Biodiversity Information Facility (GBIF), (ii) connects these data at the specimen level, and (iii) provides customized web-based apps that enable these data to be analyzed in a variety of ways.
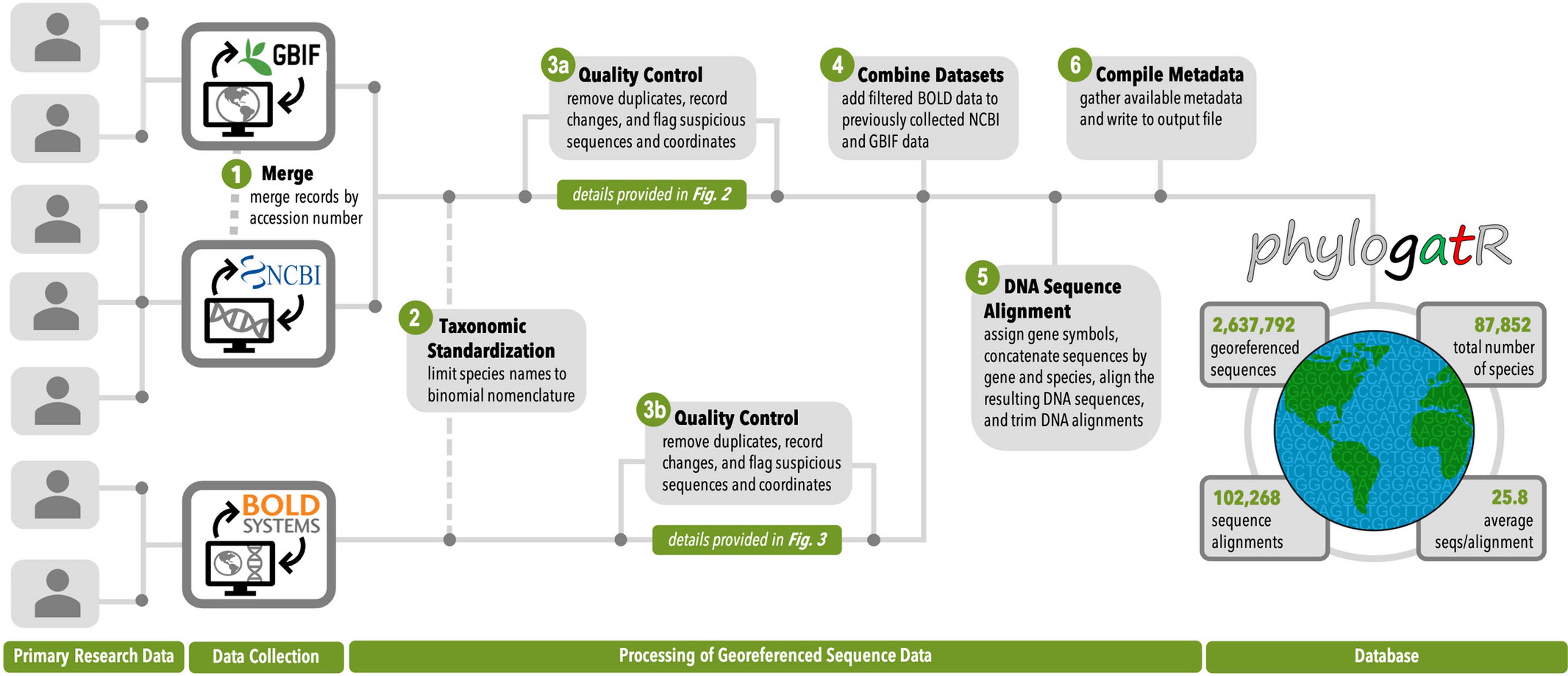 Overview of PhylogatR pipeline (from Pelletier et al. 2022)
Overview of PhylogatR pipeline (from Pelletier et al. 2022)
To ask questions or report bugs, please send a message to Phylogatr@lists.osu.edu and include the alignment name (Genus-species-gene), number(s) of the problematic accession(s), and a short description of the issue.
Submit corrections to GenBank sequence data by sending an error report to update@ncbi.nlm.nih.gov.
Submit corrections to GBIF occurrence data by clicking the speech bubble icon in the upper right corner of the GBIF website.
Project Team
- Bryan Carstens
- Tara Pelletier
- Danielle Parsons
- Sydney Decker
- Eric Franz
- Jeffrey Ohrstrom
- Shameema Ookital
- Trey Dockendorf
- Samir Mansour
The suggested citation for phylogatR is:
Tara A. Pelletier, Danielle J. Parsons, Sydney K. Decker, Stephanie Crouch, Eric Franz, Jeffrey Ohrstrom, Bryan C. Carstens. 2022. PhylogatR: Phylogeographic data aggregation and repurposing. Molecular Ecology Resources. https://doi.org/10.1111/1755-0998.13673